Simple technology makes CRISPR gene editing cheaper
Technique can quickly turn entire genomes into thousands of probes
July 23, 2015
University of California, Berkeley, researchers have discovered a much cheaper and easier way to target a hot new gene editing tool, CRISPR-Cas9, to cut or label DNA.

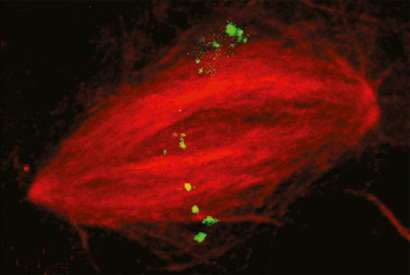
Some DNA sequences appear multiple times in the genome. Here, an RNA guide probe labels repetitive regions in the nucleus of a sperm cell from the frog Xenopus laevis.
The CRISPR-Cas9 technique, invented three years ago at UC Berkeley, has taken genomics by storm, with its ability to latch on to a very specific sequence of DNA and cut it, inactivating genes with ease. This has great promise for targeted gene therapy to cure genetic diseases, and for discovering the causes of disease.
The technology can also be tweaked to latch on without cutting, labeling DNA with a fluorescent probe that allows researchers to locate and track a gene among thousands in the nucleus of a living, dividing cell.
The newly developed technique now makes it easier to create the RNA guides that allow CRISPR-Cas9 to target DNA so precisely. In fact, for less than $100 in supplies, anyone can make tens of thousands of such precisely guided probes covering an organism’s entire genome.
The process, which they refer to as CRISPR-EATING – for “Everything Available Turned Into New Guides” – is reported in a paper to appear in the August 10 issue of the journal Developmental Cell.
As proof of principle, the researchers turned the entire genome of the common gut bacterium E. coli into a library of 40,000 RNA guides that covered 88 percent of the bacterial genome. Each RNA guide is a segment of 20 RNA base pairs: the template used by CRISPR-Cas9 as it seeks out complementary DNA to bind and cut.

The researchers created hundreds of RNA guide probes for a small region of the genome of the frog Xenopus laevis, attached them to CRISPR-Cas9 with a fluorescent label, and successfully tagged that region of DNA in a nucleus so that it can be followed in living samples.
These libraries can be employed in traditional CRISPR-Cas9 editing to target any specific DNA sequence in the genome and cut it, which is what researchers do to pin down the function of a gene: knock it out and see what bad things happen in the cell. This can help pinpoint the cause of a disease, for example. The process is called genetic screening and is done in batches: each of the thousands of probes is introduced into a single cell on a plate filled with hundreds of thousands of cells.
“We can make these libraries for a lot less money, which makes genetic screening potentially accessible in organisms less well studied,” such as those that have not yet had their genomes sequenced, said first author Andrew Lane, a UC Berkeley post-doctoral fellow.
Real-time cell monitoring
But Lane and colleague Rebecca Heald, UC Berkeley professor of molecular and cell biology, developed the technology in order to track chromosomes in real-time in living cells, in particular during cell division, a process known as mitosis. This is part of a larger project by Heald to find out what regulates the size of the nucleus and other subcellular components as organisms grow from just a few cells to many cells.
“This technology will allow us to paint a whole chromosome and look at it live and really follow it in the nucleus during the cell cycle or as it goes through developmental transitions, for example in an embryo, to see how it changes in size and structure,” Heald said.
The new technique uses standard PCR (polymerase chain reaction) to generate many short lengths of DNA from whatever segment of DNA a researcher is interested in, up to and including an entire genome. These fragments are then precisely snipped at a region called a PAM, which is critical to CRISPR binding. Simple restriction enzymes are then used to cut each piece 20 base pairs from the PAM end, generating the exact size of RNA guide that CRISPR uses in searching the genome for complementary sites. These guide RNAs are then easily incorporated into the CRISPR-Cas9 complex, yielding tens of thousands of probes for labeling or cutting DNA.
“By using the genome itself as a source for guide RNAs, their approach puts the creation of libraries that target contiguous regions in reach of almost any lab,” said Jacob Corn, managing and scientific director of the Innovative Genomics Initiative at UC Berkeley. “This could be very useful for genome imaging and certain kinds of screens, and I’m very interested to see how it enables biological discovery using Cas9 tools.”
Lane and Heald’s coauthors are Magdalena Strzelecka, Andreas Ettinger, Andrew W. Grenfell and Torsten Wittmann. This work was supported by the National Institutes of Health.
RELATED INFORMATION