$65.5 million from NIH to create brain atlas
Identifying all types of cells in the mouse brain will lead to similar atlas of human brain
October 23, 2017
UC Berkeley is partnering with the Allen Institute for Brain Science in Seattle on a $65.5 million, five-year effort to count, catalog and connect the many different cell types in the mouse brain, as a foundation for doing the same for the human brain.
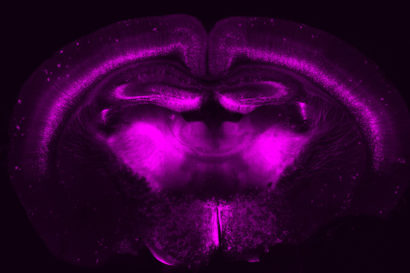
Slice of a mouse brain showing a specific set of neurons genetically tagged using the CRISPR-based gene-engineering approach developed in John Ngai’s lab with funding from the BRAIN Initiative. (David Stafford and Rebecca Chance photo)
Funded by the National Institutes of Health, the Allen Institute-led consortium represents an international team of scientists that will construct a comprehensive whole-brain atlas of cell types, essentially a parts list of the mouse brain. The effort is part of the federal government’s BRAIN Initiative, announced in 2013 with the ultimate goal of understanding brain circuits well enough to devise new therapies for diseases of the human brain and nervous system.
“Our task is to identify, classify and characterize all the cell types in the mouse brain and understand how they wire up, to serve as a basis for understanding how the human brain functions in health and disease,” said neuroscientist John Ngai, a professor of molecular and cell biology and principal investigator for the UC Berkeley team.
That is a large task, he admits, since the human brain contains about 80 billion nerve cells, called neurons, and an untold number of types of neurons.
“We’ve known about the existence of neurons for over a hundred years, but we don’t have a good handle on how many there are and what sets them apart from each other — there could be hundreds of types, there could be thousands. We now have the tools to find out,” he said.
In 2014, the NIH funded 10 pilot projects — which together comprised the BRAIN Initiative Cell Census Consortium — to explore the feasibility of various techniques for identifying different types of cells in the brain. Ngai received one of these grants. His group and others demonstrated that it is possible to identify and classify brain cells based on the genes activated in them.
When a gene is activated, or expressed, a copy is made, called messenger RNA or mRNA. The mRNA is translated by the cell’s internal machinery to construct a protein. Using a relatively new technique called single-cell transcriptomics, it is possible to find or “profile” all the mRNA transcribed in a single cell, which in turn allows scientists to group or “classify” cells based on their similar mRNA profiles.
The new project — one of nine BRAIN Initiative Cell Census Network grants announced today — brings together researchers from some of these earlier pilot projects and new colleagues from Europe and China to scale up these technologies to build a complete cell atlas of the mouse brain and reference atlases for the human brain.
“Building a comprehensive atlas of cell types requires a collaborative, multidisciplinary approach,” said Hongkui Zeng, the executive director of structured science at the Allen Institute for Brain Science and lead principal investigator of the consortium. “Joining us in this endeavor are globally recognized leaders in the fields of transcriptomics, neuroanatomy, neurophysiology, computational biology and microscopy. Our collective efforts will accelerate the entire community’s progress in decoding brain function and impact everything from disease research to artificial intelligence.”
The Allen Institute consortium will analyze the transcriptomes of single cells plucked from throughout the brain, and work with statisticians to decide how to use this mRNA profile to lump cells together and relate that to their shape, their function and their connectivity to other cells in the brain.
“If you know all this information you pretty much have a comprehensive description of what a cell is and a way of understanding what it does in processing information in the mammalian brain,” Ngai said. “It’s exciting that with this single-cell approach, we are able to see fine distinctions between cells we had not been able to see before and had no way of knowing how to look for, just by letting the cells tell us by a show of hands, or of genes, which ones are different from another.”
Once cell types are identifiable by their gene-expression profiles, it will be possible to target and study them, he said. The UC Berkeley team plans to use CRISPR-Cas9 technology to generate transgenic mice based on specific mRNA profiles that researchers anywhere can use to study the properties of the newly discovered cell types.
In addition to Ngai and Zeng, the principal investigators are David Anderson and Lior Pachter of the California Institute of Technology, Xiaowei Zhuang of Harvard University and Andreas Tolias of Baylor College of Medicine. The UC Berkeley group’s co-investigators are Hillel Adesnik and Dirk Hockemeyer from the Department of Molecular and Cell Biology, Sandrine Dudoit from the Department of Biostatistics, Elizabeth Purdom from the Department of Statistics and statistician Davide Risso from Weill Cornell Medicine in New York.
The project also includes co-investigators from the Massachusetts Institute of Technology, Huazhong University of Science and Technology, Karolinska Institute, University of Tubingen and collaborators from the Howard Hughes Medical Institute Janelia Research Campus.
RELATED INFORMATION