Freeze-frame microscopy captures molecule’s ‘lock-and-load’ on DNA
Scientists obtain clearer picture of key machinery that transcribes DNA, thanks to cryo-electron microscopy
November 16, 2018
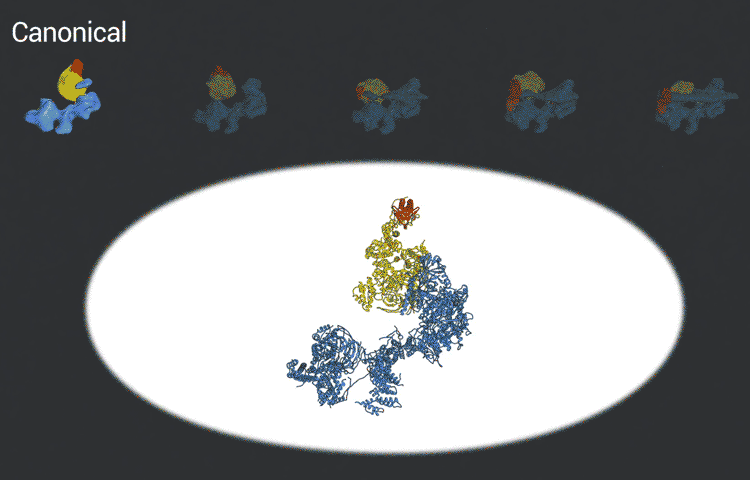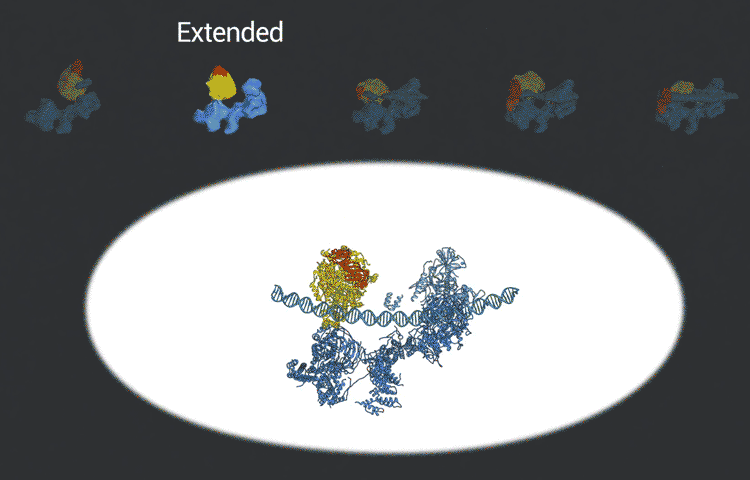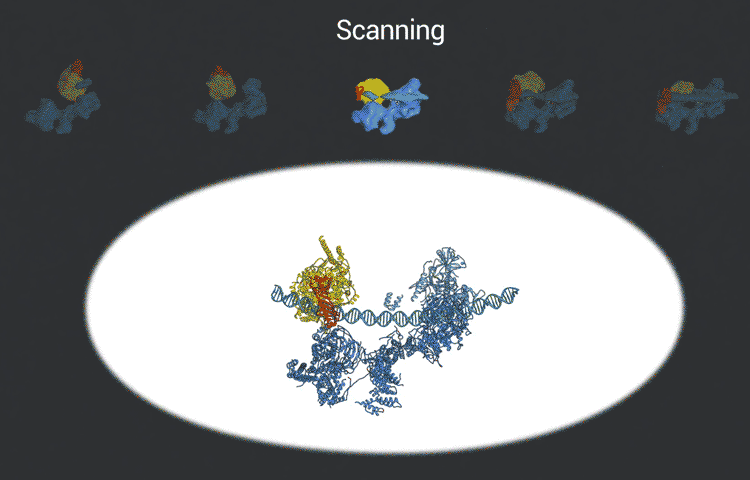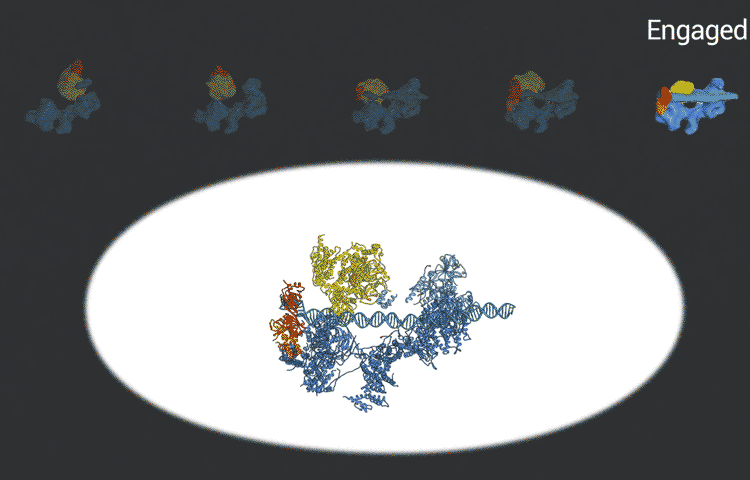
The transcription factor IID complex locks onto DNA, checks it’s in the right place and then recruits other proteins to start transcribing DNA into RNA. New advances in cryo-EM allowed researchers to define five distinct conformations of TFIID as it locks and loads. (Eva Nogales lab)
Pushing the limits of cryo-electron microscopy, University of California, Berkeley, scientists have captured freeze-frames of the changing shape of a huge molecule, one of the body’s key molecular machines, as it locks onto DNA and loads the machinery for reading the genetic code.
The molecule, called transcription factor IID, is critical to transcribing genes into RNA that will later be used as blueprints to make proteins. Because of its many moving parts and large size, however, TFIID’s 3D structure has been hard to capture: the moving parts become a blur.
Cryo-EM, an imaging technique whose discoverers garnered the 2017 Nobel Prize in Chemistry, is the only way to obtain a snapshot of bulky, floppy structures like this. High-resolution structural information is essential for understanding how TFIID translates the operating instructions in our genome and how it sometimes goes haywire.
The new, more detailed snapshots of the molecule’s moving parts could help drug designers create drugs that interfere with the molecule’s structural changes in order to tweak the expression of a gene that is causing disease.
“These structures give you the potential for rationally designing small molecules that will disturb the normal function, because now we don’t have just a single structure, we have many structures, which is even more powerful because we can target the motion that we are seeing right now,” said Eva Nogales, a UC Berkeley professor of molecular and cell biology and a faculty scientist at Lawrence Berkeley National Laboratory.
Nogales and her colleagues, most prominently UC Berkeley graduate students Avinash Patel and Robert Louder, posted their findings online this week in advance of print publication in the journal Science.
“Where you stick that drug and how you make that drug operate is highly dependent on the transient nature of these structures, and that is something we have come to realize only very recently,” said Robert Tjian, a UC Berkeley professor of molecular and cell biology who discovered TFIID and has worked on the molecule for much of his career, though he was not a co-author of the new paper. “Because these molecules are moving around and their structures are so complicated, conventional drug discovery would never reveal what is going on. Eva’s structures are going to change that. This has the potential to open up the universe of druggable targets.”
Freezing molecules
TFIID is an agglomeration of more than a dozen distinct proteins that homes in on a promoter – a region of DNA that controls the transcription of a nearby gene – and tests the sequence to make sure it has landed at the right spot. Once this is confirmed, it opens up to recruit dozens of other proteins that then start ratcheting along the gene, using the DNA sequence as a template to create a complementary sequence of RNA, called messenger RNA. This then wends its way out of the nucleus into the body of the cell, where it is translated by other molecular machines into protein.
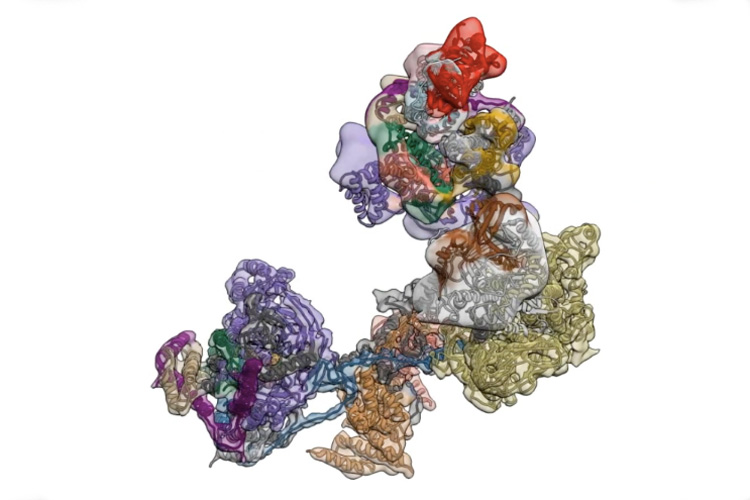
TFIID is a complex of proteins that initiates transcription of the body’s operating instructions encoded in DNA.
“TFIID is probably the hardest structure anybody has tried to solve, because it is massive and it is highly flexible,” said Tjian. “The only way you can see these very flexible structures is by cryo-EM, and Eva can now freeze all these different flexible states and describe the movement.”
Cryo-EM involves freezing a drop containing millions of copies of a molecule, in every imaginable orientation, and using an electron microscope to determine the structure by combining images to define the 3D shape. Because TFIID has many moving parts as it binds to DNA and gets ready to transcribe a gene, averaging all the frozen positions produces a blurred image.
Nogales compares previous efforts at imaging TFIID, including her own attempts going back nearly 20 years, to taking a photograph of a soccer team on the field and averaging the motions of all the players. The result is a generic human torso with blurry head and limbs.
Imaging the soccer field
Now, thanks to more than two years of intensive work by Patel and Louder, it’s possible to capture higher resolution images that are akin to distinguishing a striker’s leg kick from a goalie’s hand block and a fullback’s header.
“Imagine you have an image of the 22 soccer players on the field, and you were to combine them into a single unit that you would call the ‘generic soccer player,’” she said. “It will look like a blurry picture — you could barely see that it’s a humanoid form and that there is some kind of motion, but you wouldn’t realize that a difference exists between the players.”
The improved pictures are a result of better detectors developed originally by colleagues at Berkeley Lab, and steadily improving computer algorithms to analyze the huge amounts of data collected by the detectors. This helped Nogales and her team to define five distinct structures of the TFIID molecule.
“They span the whole binding sequence: before binding to the DNA, initial binding to the promoter, subsequent binding after it double checks that this is the right place, and the final state,” Nogales said.
She and her colleagues continue to push the limits of cryo-EM, hoping to determine the 3D structure of TFIID after other transcription proteins land on it to complete the transcription process.
Among Nogales’ co-authors are Avinash Patel and Robert Louder of UC Berkeley and Berkeley Lab and Basil Greber of Berkeley Lab. The work was funded by the National Institute of General Medical Sciences (R01-GM63072, R01-GM053451, P50-GM076547) and National Cancer Institute (R21-CA175849).
RELATED INFORMATION
- Structure of human TFIID and mechanism of TBP loading onto promoter DNA (Science First Release)
- Unlocking the Secrets of Gene Expression (March 2016)
- UC Berkeley, LBNL scientists snap first 3-D pictures of the “heart” of the transcription machine (Dec. 1999)
- Eva Nogales lab website
- Robert Tjian lab website